The Rowley Lab for 3D Chromatin Organization
Genomes are intricately folded within the 3D nucleus, providing the context for numerous biological functions. Our research uncovers governing principles of 3D genome folding using a combined bioinformatics and wet-lab experimental approach. We work to better understand the relationships between chromatin, transcription, genome folding, and disease.
Methods
We primarily use genomic methods, including derivations of Hi-C, ATAC-seq, CUT&RUN, and RNA-seq to study features of chromtin organization. We are particularly interested in innovating both the methodolgy and the analysis that is commonly used to study chromatin architecture in order to understand genome organization at sub-kilobase resolution.
Software Development
Part of our work is to develop novel algorithms and easy to use tools for the analysis of genomics data, particuraly data obatined by Hi-C or its derivatives.
Model Systems
We are advocates of using model organisms, which are essential for progress in genetics and have been instrumental in discovering principles of genome organization. We have worked in plants, worms, and flies, each of which has similar and distinct ways of organizing their genome. Understanding these principles is helping to uncover the basic rules of chromatin architecture.
software
Our tools are open-source and versatile. We hope they facilitate your analysis of 3D genome architecture.
CRUSH (coming soon...)
Compartment Refinement for Ultraprecise Stratification of Hi-C (CRUSH) enables sub-kilobase resolution compartment identification from 1/100,000 the reads as well as versatile identification of genome-wide interaction preferences for any feature.
Developer: Achyuth Kalluchi
HiCrayon
HiCrayon colors 2D interactions by 1D genomic data. Web app and downloadable versions are available and are particularly useful for striking visuals and comparison of protein occupancy and long-range chromatin interactions.
Get the Full Version on GitHub!
Developer: Ben Nolan
JukeBox (coming soon...)
JukeBox measures noise within a Hi-C map and can be used to evaluate the quality of maps, to predict the return on additional sequencing, or to denote genomic regions of high noise that should be omitted from downstream analysis.
Developer: Tim Reznicek
RIPPLE (coming soon...)
Radial Identification of Proximal Propagation within Looped Entities (RIPPLE) identifies the central peak of loop signal and measures the propogation of interactions into proximal loci.
Developer: Hannah Harris
SIP
Significant Interaction Peak caller (SIP) is a tool that use image processing to identify and analyze loops that appear as high intensity signal in Hi-C maps.
Developer: Axel Poulet
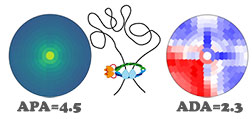
SIPMeta
SIPMeta is used to visualize averages of loops and includes a novel "bullseye" transformation to eliminate visualization biases present in square plots. It also performs aggregate peak analysis (APA) and aggregate domain analysis (ADA).
Developer: Axel Poulet
JRowleyLab GitHub
Visit our github page for more! Links to POSSUMM, the NIH/NIGMS Sandbox, and other tools/resources that we have collaborated on or are under development.
Bioinformatics Education
Bioinformatics analysis of 'omics data has become an essential part of biological reasearch. We are committed to educating researchers in analyzing various types of genomic data.
View the Bioinformatics & Systems Biology (BISB) PhD program at the University of Nebraska Medical Center.
We contribute to cloud-based education as part of the NIH/NIGMS Sandbox tutorials.
We also provide assistance with DECODE, an NIH funded T32 program to improve training in 'Big Data'.
People
Meet the members of the Rowley Lab!




Jordan Rowley, PhD
Principal InvestigatorAssociate Professor
Dep. Genetics, Cell Biology, and Anatomy
Director of the BISB program at UNMC

Chris Cummings, MD/PhD
InvestigatorAssistant Professor
Childrens Health Research Institute

Hannah Harris, PhD
Postdoc
Achyuth Kalluchi
PhD Student
Tim Reznicek
PhD Student
Ben Nolan
PhD Student
Joynob Akter Puspo
PhD Student
Iesha Bryant
PhD Student
Rebecca Wright
PhD Student
You?
Contact us- Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet. 7 : e1002120. 2011
- Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 26 : 1825-36. 2012
- Zheng Q, Rowley MJ, Böhmdorfer G, Sandhu D, Gregory BD, Wierzbicki AT. RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J. 73 : 179-89. 2013
- Zhu Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell. 49 : 298-309. 2013
- Rowley MJ, Böhmdorfer G, Wierzbicki AT. Analysis of long non-coding RNAs produced by a specialized RNA polymerase in Arabidopsis thaliana. Methods. 63 : 160-9. 2013
- Böhmdorfer G, Rowley MJ, Kucinski J, Zhu Y, Amies I, Wierzbicki AT. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J. 79 : 181-91. 2014
- Rowley MJ, Corces VG. The three-dimensional genome: principles and roles of long-distance interactions. Curr. Opin. Cell Biol. 40 : 8-14. 2016
- Rowley MJ, Corces VG. Minute-Made Data Analysis: Tools for Rapid Interrogation of Hi-C Contacts. Mol. Cell. 64 : 9-11. 2016
- Böhmdorfer G, Sethuraman S, Rowley MJ, Krzyszton M, Rothi MH, Bouzit L, Wierzbicki AT. Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. Elife. 5 : 2016
- Rowley MJ, Corces VG. Capturing native interactions: intrinsic methods to study chromatin conformation. Mol. Syst. Biol. 12 : 897. 2016
- Ye R, Chen Z, Lian B, Rowley MJ, Xia N, Chai J, Li Y, He XJ, Wierzbicki AT, Qi Y. A Dicer-Independent Route for Biogenesis of siRNAs that Direct DNA Methylation in Arabidopsis. Mol. Cell. 61 : 222-35. 2016
- Cubeñas-Potts C, Rowley MJ, Lyu X, Li G, Lei EP, Corces VG. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 45 : 1714-1730. 2017
- Gómez-Díaz E, Yerbanga RS, Lefèvre T, Cohuet A, Rowley MJ, Ouedraogo JB, Corces VG. Epigenetic regulation of Plasmodium falciparum clonally variant gene expression during development in Anopheles gambiae. Sci Rep. 7 : 40655. 2017
- Rowley MJ, Rothi MH, Böhmdorfer G, Kucinski J, Wierzbicki AT. Long-range control of gene expression via RNA-directed DNA methylation. PLoS Genet. 13 : e1006749. 2017
- Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, Corces VG. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol. Cell. 67 : 837-852.e7. 2017
- Ando-Kuri M, Rivera ISM, Rowley MJ, Corces VG. Analysis of Chromatin Interactions Mediated by Specific Architectural Proteins in Drosophila Cells. Methods Mol. Biol. 1766 : 239-256. 2018
- Lyu X, Rowley MJ, Corces VG. Architectural Proteins and Pluripotency Factors Cooperate to Orchestrate the Transcriptional Response of hESCs to Temperature Stress. Mol. Cell. 71 : 940-955.e7. 2018
- Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19 : 789-800. 2018
- Ricci WA, Lu Z, Ji L, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejía-Guerra MK, Colomé-Tatché M, Johannes F, Rowley MJ, Corces VG, Zhai J, Scanlon MJ, Buckler ES, Gallavotti A, Springer NM, Schmitz RJ, Zhang X. Widespread long-range cis-regulatory elements in the maize genome. Nat Plants. 5 : 1237-1249. 2019
- Rowley MJ, Lyu X, Rana V, Ando-Kuri M, Karns R, Bosco G, Corces VG. Condensin II Counteracts Cohesin and RNA Polymerase II in the Establishment of 3D Chromatin Organization. Cell Rep. 26 : 2890-2903.e3. 2019
- Jung YH, Kremsky I, Gold HB, Rowley MJ, Punyawai K, Buonanotte A, Lyu X, Bixler BJ, Chan AWS, Corces VG. Maintenance of CTCF- and Transcription Factor-Mediated Interactions from the Gametes to the Early Mouse Embryo. Mol. Cell. 75 : 154-171.e5. 2019
- Gutierrez-Perez I, Rowley MJ, Lyu X, Valadez-Graham V, Vallejo DM, Ballesta-Illan E, Lopez-Atalaya JP, Kremsky I, Caparros E, Corces VG, Dominguez M. Ecdysone-Induced 3D Chromatin Reorganization Involves Active Enhancers Bound by Pipsqueak and Polycomb. Cell Rep. 28 : 2715-2727.e5. 2019
- Fernandez-Albert J, Lipinski M, Lopez-Cascales MT, Rowley MJ, Martin-Gonzalez AM, Del Blanco B, Corces VG, Barco A. Immediate and deferred epigenomic signatures of in vivo neuronal activation in mouse hippocampus. Nat. Neurosci. 22 : 1718-1730. 2019
- Rowley MJ; Poulet A; Nichols MH; Bixler, BJ; Sanborn AL; Brouhard AH; Hermetz K; Linsenbaum H; Csankovszki G; Aiden E; Corces VG. Analysis of Hi-C data using SIP effectively identifies loops in organisms from C. elegans to mammals. Genome research. 2020 Mar; Vol. 30 (3): p. 447-458. 1088-9051
- Kucinski J, Chamera S, Kmera A, Rowley MJ, Fujii S, Khurana P, Nowotny M, Wierzbicki AT. Evolutionary history and activity of RNase H1-like proteins in Arabidopsis thaliana. Plant Cell Physiol. 2020
- Engstrom AK, Walker AC, Moudgal RA, Myrick DA, Kyle SM, Bai Y, Rowley MJ, Katz DJ. The inhibition of LSD1 via sequestration contributes to tau-mediated neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America. 117 : 29133-29143. 2020
- Rocks D; Shukla M; Finneman; Kalluchi A; Rowley MJ; Kundakovic M. Sex-specific multi-level 3D genome dynamics in the mouse brain. Nature Communications. 2022
- Mirza S, Kalluchi A, Raza M, Saleem I, Mohapatra B, Pal D, Ouellette MM, Qiu F, Yu L, Lobanov A, Zheng ZM, Zhang Y, Alsaleem MA, Rakha EA, Band H, Rowley MJ, Band V. Ecdysoneless Protein Regulates Viral and Cellular mRNA Splicing to Promote Cervical Oncogenesis. Molecular Cancer Research. 20 : 305-318. 2022.
- Cummings CT, Rowley MJ. Implications of Dosage Deficiencies in CTCF and Cohesin on Genome Organization, Gene Expression, and Human Neurodevelopment. Genes. 13 : 2022
- Dale GA, Wilkins DJ, Rowley J, Scharer CD, Tipton CM, Hom J, Boss JM, Corces V, Sanz I, Jacob J. Somatic Diversification of Rearranged Antibody Gene Segments by Intra- and Interchromosomal Templated Mutagenesis. Journal of immunology (Baltimore, Md. : 1950). 208 : 2141-2153. 2022
- Metz EP, Wilder PJ, Popay TM, Wang J, Liu Q, Kalluchi A, Rowley MJ, Tansey WP, Rizzino A. Elevating SOX2 Downregulates MYC through a SOX2:MYC Signaling Axis and Induces a Slowly Cycling Proliferative State in Human Tumor Cells. Cancers. 14 : 2022
- Kalluchi A; Harris HL; Reznicek TE; Rowley MJ. Considerations and Caveats for Analyzing Chromatin Compartments. Frontiers in Molecular Bioscience. 2023
- Zhou Y, Dogiparthi VR, Ray S, Schaefer M, Harris HL, Rowley MJ, Hewitt KJ. Defining A Cohort of Anemia-Activated Cis-Elements Reveals a Mechanism Promoting Erythroid Precursor Function. Blood advances. 2023
- Chakraborty S, Bhat AM, Mushtaq I, Luan H, Kalluchi A, Mirza S, Storck MD, Chaturvedi N, Lopez-Guerreo JA, Llombart-Bosch A, Machado I, Scotlandi K, Meza JL, Ghosal G, Coulter DW, Rowley MJ, Band V, Mohapatra BC, Band H. EHD1-dependent traffic of IGF-1 receptor to the cell surface is essential for Ewing sarcoma tumorigenesis and metastasis. Communications Biology 2023
- Harris HL, Gu H, Olshansky M, Wang A, Farabella I, Eliaz Y, Kalluchi A, Krishna A, Jacobs M, Cauer G, Pham M, Rao S, Dudchenko O, Omer A, Mohajeri K, Kim S, Nichols MH, Davis E, Gkountaroulis D, Udupa D, Aiden A, Corces VG, Phanstiel D, Noble W, Nir G, Di Pierro M, Seo J, Talkowski M, Aiden E, Rowley MJ. Chromatin alternates between A and B compartments at kilobase scale for subgenic organization. Nature Communications. 2023
- Puerto M, Shukla M, Bujosa P, Perez-Roldan J, Torras-Llort M, Tamirisa S, Carbonell A, Sole C, Puspo JA, Cummings CT, de Nadal E, Posas F, Azorin F, Rowley MJ. The zinc-finger protein Z4 cooperates with condensin II to regulate somatic chromosome pairing and 3D chromatin organization. Nucleic Acids Res. 2024
- Lyu X, Rowley MJ, Kulik MJ, Dalton S, Corces VG. Regulation of CTCF loop formation during pancreatic cell differentiation. Nat Comm. 2023.
- Nolan B, Harris HL, Kalluchi A, Reznicek TE, Cummings CT, Rowley MJ. HiCrayon reveals distinct layers of multi-state 3D chromatin organization. bioRxiv 2024.
- Clark KL, Shukla M, George JW, Gustin S, Rowley MJ, Davis JS. An environmentally relevant mixture of Perfluoroalkyl Substances (PFAS) impacts proliferation, steroid hormone synthesis, and gene transcription in primary human granulosa cells. Toxicol Sci. 2024.
- Harris HL, Rowley MJ. Mechanistic drivers of chromatin organization into compartments. Curr Opin Genet Dev. 2024.
- Bhattacharya S, Harris HL, Islam R, Bodas S, Polavaram N, Mishra J, Das D, Seshacharyulu P, Kalluchi A, Pal A, Kohli M, Lele SM, Muders M, Batra SK, Ghosh PM, Datta K, Rowley MJ, Dutta S. Understanding the function of Pax5 in development of docetaxel-resistant neuroendocrine-like prostate cancers. Cell Death Dis. 2024.
- Veerappa AM, Rowley MJ, Maggio A, Beaudry L, Hawkins D, Kim A, Sethi S, Sorgen PL, Guda C. CloudATAC: a cloud-based framework for ATAC-Seq data analysis. Brief Bioinform. 2024.
- Sandoval-Velasco M, Dudchenko O, Rodríguez JA, Pérez Estrada C, Dehasque M, Fontsere C, Mak SST, Khan R, Contessoto VG, Oliveira Junior AB, Kalluchi A, Zubillaga Herrera BJ, Jeong J, Roy RP, Christopher I, Weisz D, Omer AD, Batra SS, Shamim MS, Durand NC, O'Connell B, Roca AL, Plikus MV, Kusliy MA, Romanenko SA, Lemskaya NA, Serdyukova NA, Modina SA, Perelman PL, Kizilova EA, Baiborodin SI, Rubtsov NB, Machol G, Rath K, Mahajan R, Kaur P, Gnirke A, Garcia-Treviño I, Coke R, Flanagan JP, Pletch K, Ruiz-Herrera A, Plotnikov V, Pavlov IS, Pavlova NI, Protopopov AV, Di Pierro M, Graphodatsky AS, Lander ES, Rowley MJ, Wolynes PG, Onuchic JN, Dalén L, Marti-Renom MA, Gilbert MTP, Aiden EL. Three-dimensional genome architecture persists in a 52,000-year-old woolly mammoth skin sample. Cell. 2024.
- George JW, Cancino RA, Griffin Miller JL, Qiu F, Lin Q, Rowley MJ, Chennathukuzhi VM, Davis JS. Characterization of m6A Modifiers and RNA Modifications in Uterine Fibroids. Endocrinology. 2024.
- Smith AL, Skupa SA, Eiken AP, Reznicek TE, Schmitz E, Williams N, Moore DY, D'Angelo CR, Kallam A, Lunning MA, Bociek RG, Vose JM, Mohamed E, Mahr AR, Denton PW, Powell B, Bollag G, Rowley MJ, El-Gamal D. BET inhibition reforms the immune microenvironment and alleviates T cell dysfunction in chronic lymphocytic leukemia. JCI Insight. 2024
- Nolan B, Reznicek TE, Cummings CT, Rowley MJ. The chromatin tapestry as a framework for neurodevelopment. Genome Res. 2024.
- Bhat AM, Mohapatra BC, Luan H, Mushtaq I, Chakraborty S, Kumar S, Wu W, Nolan B, Dutta S, Storck MD, Schott M, Meza JL, Lele SM, Lin MF, Cook LM, Corey E, Morrissey C, Coulter DW, Rowley MJ, Natarajan A, Datta K, Band V, Band H. GD2 and its biosynthetic enzyme GD3 synthase promote tumorigenesis in prostate cancer by regulating cancer stem cell behavior. Sci Rep. 2024.
- Smith AL, Skupa SA, Eiken AP, Reznicek TE, Schmitz E, Williams N, Moore DY, D'Angelo CR, Kallam A, Lunning MA, Bociek RG, Vose JM, Mohamed E, Mahr AR, Denton PW, Powell B, Bollag G, Rowley MJ, El-Gamal D. BET inhibition reforms the immune microenvironment and alleviates T cell dysfunction in chronic lymphocytic leukemia. JCI Insight. 2024 May.
- Eiken AP, Smith AL, Skupa SA, Schmitz E, Rana S, Singh S, Kumar S, Mallareddy JR, de Cubas AA, Krishna A, Kalluchi A, Rowley MJ, D'Angelo CR, Lunning MA, Bociek RG, Vose JM, Natarajan A, El-Gamal D. Novel Spirocyclic Dimer, SpiD3, Targets Chronic Lymphocytic Leukemia Survival Pathways with Potent Preclinical Effects. Cancer Res Commun. 2024.
- Zheng S, Thakkar N, Harris HL, Liu S, Zhang M, Gerstein M, Aiden EL, Rowley MJ, Noble WS, Gürsoy G, Singh R. Predicting A/B compartments from histone modifications using deep learning. iScience. 2024.
Contact
Department of Genetics, Cell Biology & Anatomy
Omaha, NE 68106
jordan.rowley@unmc.edu
@MJordanRowley